A web server of contig scaffolding using algebraic rearrangements
A web server of contig scaffolding using algebraic rearrangements
CSAR-web is a web server that can efficiently and accurately scaffold (i.e., order and orient) the contigs of a target draft genome based on a complete or incomplete reference genome of a related organism.
CSAR-web provides a web interface (see Figure 1) that is intuitive and easy to operate. For convenience, the user can choose one of the examples (1) we prepared in advance for running CSAR-web, or submit a job according to the procedures described below.
The user can click "Help" tab page (8) for the details of how to run CSAR-web.
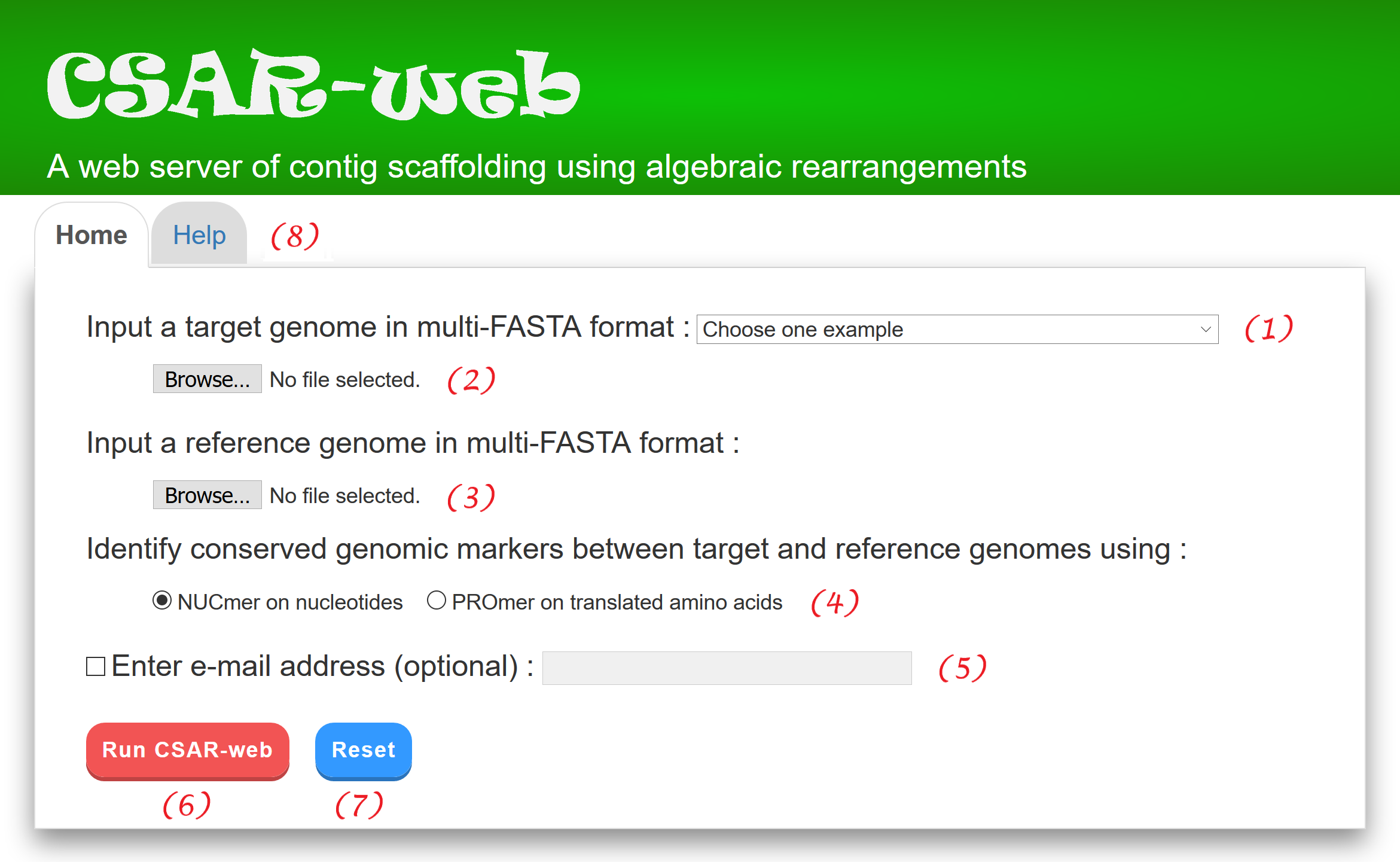
Figure 1:
User interface of CSAR-web.
CSAR-web outputs its scaffolding results in four tab pages: (1) input data & parameters, (2) dotplot validation, (3) scaffolds of target, and (4) scaffolds of reference. In the "Input data & parameters" page (see Figure 2 for an example), CSAR-web shows the information of input target and reference genomes, the user-specified method (either NUCmer or PROmer) for identifying their conserved genomic markers, and a dotplot for the visual inspection of identified genomic markers before scaffolding. In the dotplot, the target and reference genomes are plotted on the y and x axes, respectively, and their contigs and scaffolds are separated by horizontal or vertical dashed lines. In addition, forward and reverse matched sequence regions are displayed in red and blue lines, respectively, and the beginning and end of each line are represented by two unfilled points. Note that the users have an option to sort the input contigs of the target genome according to their sizes by using a toggle switch. The users can also zoom in or out on a particular region of the dotplot by clicking the ``Zoom in'' or ``Zoom out'' button, respectively (or simply by scrolling the mouse wheel over the dotplot). Furthermore, the users can show or hide the numbers of contigs, which are generated randomly in a format that begins with three-letter prefix (CTG) followed by an underscore (_) and at least three digits (e.g., CTG_001), by using a toggle switch.
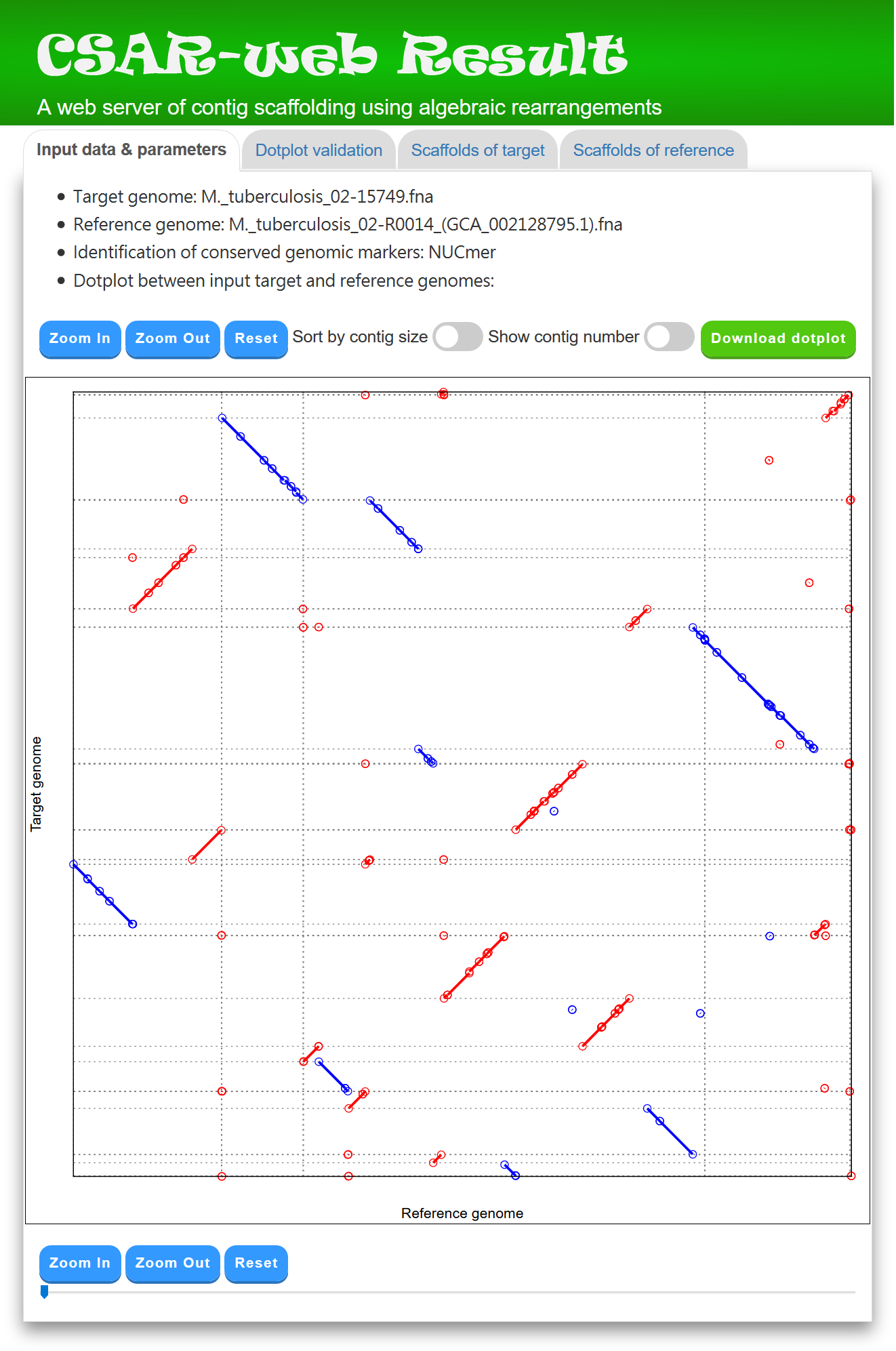
Figure 2: A display of the "Input data & parameters" tab page.
In the "Dotplot validation" page, CSAR-web displays its total running time, as well as its scaffolding result by a dotplot between the scaffolds of target and reference genomes (see Figure 3 for an example). The scaffolds of the target genome generated by CSAR-web are numbered randomly and the format of their scaffolding numbers begins with three-letter prefix (SCF) followed by an underscore (_) and at least three digits (e.g., SCF_001). In principle, if the contigs of the target genome are perfectly scaffolded according to the reference genome, then the matched regions in the dotplot would go from the bottom left to the top right (as shown in Figure 3) or go from the top left to the bottom right. The dotplot display of the scaffolding result is convenient for the user to visually validate whether the contigs of the target genome are properly scaffolded according to the reference genome. The dotplot is zoomable and the numbers of its contigs and scaffolds can be shown or hidden by using a toggle switch. In addition, the user can download a copy of the dotplot in scalar vector graphics (SVG) format, which can be opened in many Web browsers (e.g., Mozilla Firefox, Google Chrome, Apple Safari, Microsoft Internet Explorer and Microsoft Edge) and used to create a publication-quality figure, by clicking the "Download dotplot" button.
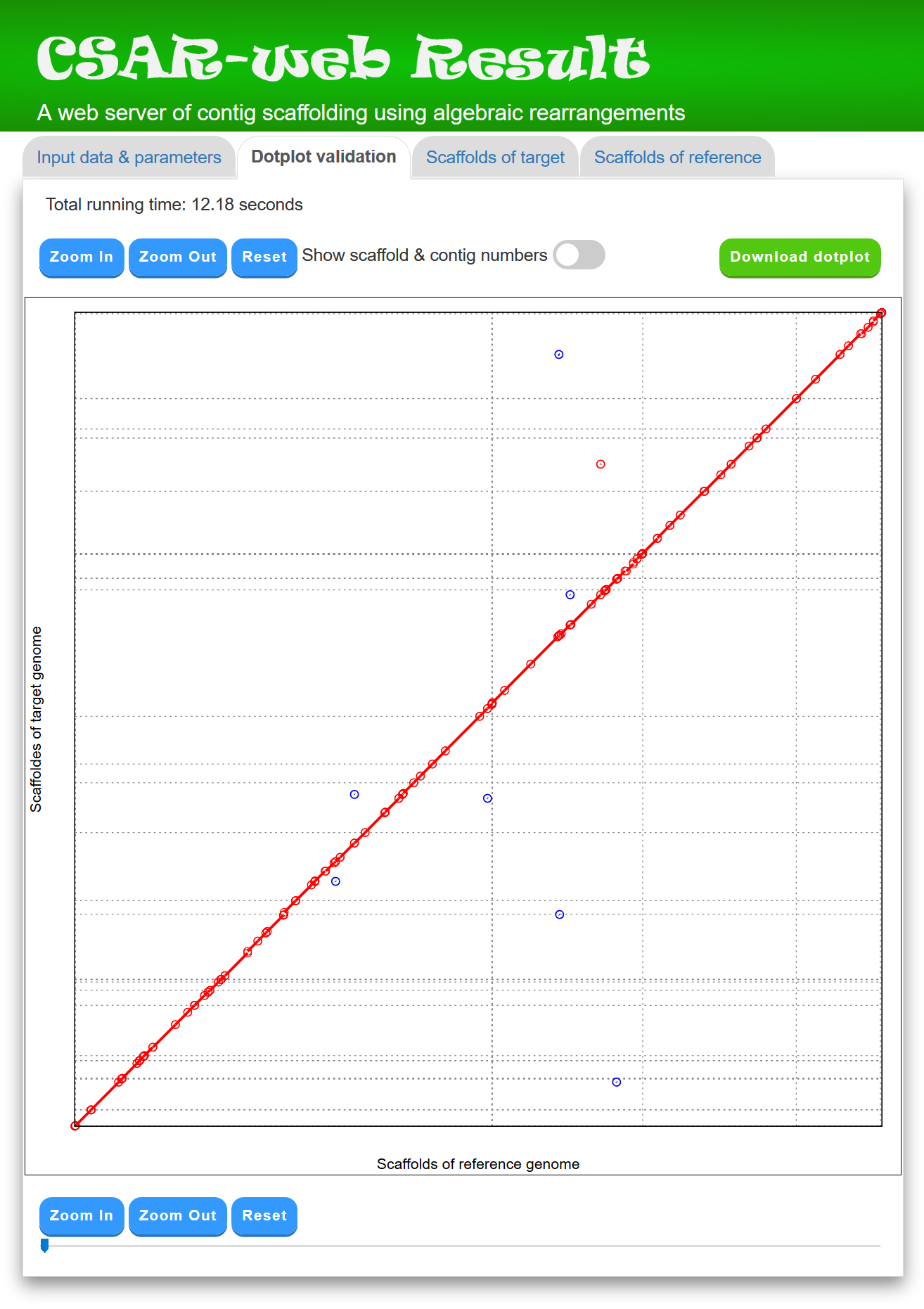
Figure 3: A display of the "Dotplot validation" tab page.
In the "Scaffolds of target" page, CSAR-web displays its scaffolding result in tabular format (see Figure 4 for an example) for the purpose of allowing the user to view the generated scaffolds in detail. The scaffolds in the table are sorted according to their sizes, which equals to the sum of contig sizes. The contigs of each generated scaffold, along with their orientation (0 standing for forward and 1 for reverse), sequence and length, are listed in a table according to their order in the scaffold. For downstream analyses, the users can download the scaffolds of target genome either in a tab-delimited text format or a comma-delimited CSV format by clicking the "Download scaffolds (.txt)" or "Download scaffolds (.csv)" button, respectively. In addition, the users can download the scaffold sequences in the text format by clicking the Download sequences button, where contig sequences in the same scaffold are separated by 100 Ns.
In the "Scaffolds of reference" page, CSAR-web displays the scaffold table for the reference genome. Note that when the input reference genome is a draft genome, its contigs are scaffolded by CSAR-web using the input target genome as reference.
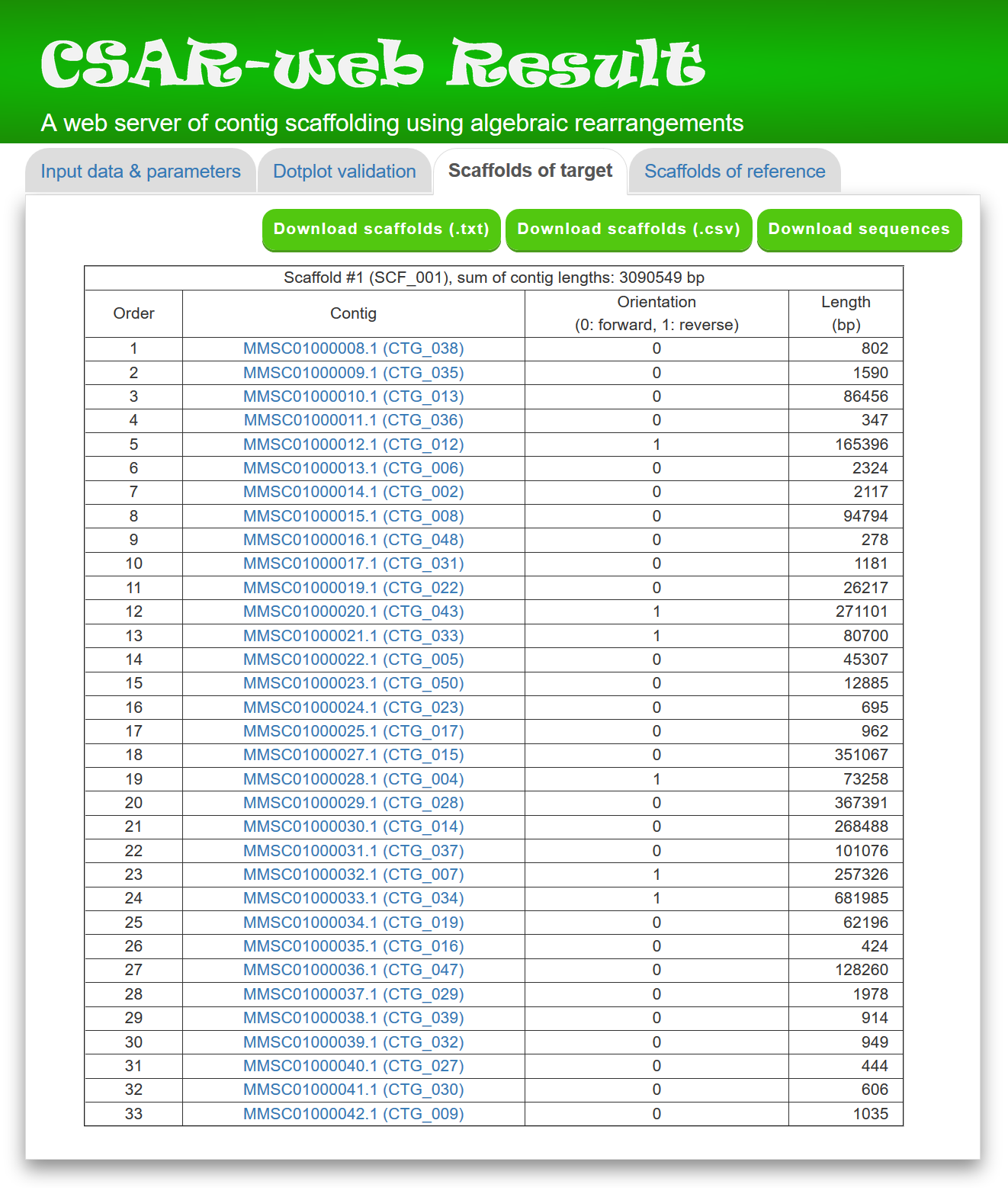
Figure 4: A partial display of the "Scaffolds of target" tab page.